Contents
Run AFQ analysis for 2 groups (patients and controls)
[AFQbase AFQdata] = AFQ_directories;
sub_dirs = {[AFQdata '/patient_01/dti30'], [AFQdata '/patient_02/dti30']...
[AFQdata '/patient_03/dti30'], [AFQdata '/control_01/dti30']...
[AFQdata '/control_02/dti30'], [AFQdata '/control_03/dti30']};
sub_group = [1, 1, 1, 0, 0, 0];
afq = AFQ_Create('run_mode','test', 'sub_dirs', sub_dirs, 'sub_group',...
sub_group, 'showfigs',false);
[afq, patient_data, control_data, norms, abn, abnTracts] = AFQ_run(sub_dirs, sub_group, afq);
Whole-brain tractography was already done for subject /Users/jyeatman/git/AFQ/data/patient_01/dti30
Fiber tract segmentation was already done for subject /Users/jyeatman/git/AFQ/data/patient_01/dti30
Fiber tract cleaning was already done for subject /Users/jyeatman/git/AFQ/data/patient_01/dti30
Computing Tract Profiles for subject /Users/jyeatman/git/AFQ/data/patient_01/dti30
Whole-brain tractography was already done for subject /Users/jyeatman/git/AFQ/data/patient_02/dti30
Fiber tract segmentation was already done for subject /Users/jyeatman/git/AFQ/data/patient_02/dti30
Fiber tract cleaning was already done for subject /Users/jyeatman/git/AFQ/data/patient_02/dti30
Computing Tract Profiles for subject /Users/jyeatman/git/AFQ/data/patient_02/dti30
Whole-brain tractography was already done for subject /Users/jyeatman/git/AFQ/data/patient_03/dti30
Fiber tract segmentation was already done for subject /Users/jyeatman/git/AFQ/data/patient_03/dti30
Fiber tract cleaning was already done for subject /Users/jyeatman/git/AFQ/data/patient_03/dti30
Computing Tract Profiles for subject /Users/jyeatman/git/AFQ/data/patient_03/dti30
Whole-brain tractography was already done for subject /Users/jyeatman/git/AFQ/data/control_01/dti30You chose to recompute ROIs
Fibers that get as close to the ROIs as 2mm will become candidates for the Mori Groups
Smoothing by 0 & 8mm..
Coarse Affine Registration..
Fine Affine Registration..
3D CT Norm...
iteration 1: FWHM = 13.31 Var = 24.3369
iteration 2: FWHM = 10.26 Var = 2.3268
iteration 3: FWHM = 9.9 Var = 1.82368
iteration 4: FWHM = 9.811 Var = 1.68895
iteration 5: FWHM = 9.746 Var = 1.62451
iteration 6: FWHM = 9.729 Var = 1.60517
iteration 7: FWHM = 9.714 Var = 1.59288
iteration 8: FWHM = 9.708 Var = 1.5878
iteration 9: FWHM = 9.7 Var = 1.58208
iteration 10: FWHM = 9.701 Var = 1.58178
iteration 11: FWHM = 9.698 Var = 1.58025
iteration 12: FWHM = 9.698 Var = 1.57987
iteration 13: FWHM = 9.697 Var = 1.57926
iteration 14: FWHM = 9.697 Var = 1.57926
iteration 15: FWHM = 9.696 Var = 1.57881
iteration 16: FWHM = 9.697 Var = 1.57881
Computing inverse deformation...
dtiCleanFibers: Keeping 32549 out of 32704 fibers.
dtiSplitInterhemisphericFibers: Splitting every fiber below Z=-10
Fiber tract cleaning was already done for subject /Users/jyeatman/git/AFQ/data/control_01/dti30
Computing Tract Profiles for subject /Users/jyeatman/git/AFQ/data/control_01/dti30
Whole-brain tractography was already done for subject /Users/jyeatman/git/AFQ/data/control_02/dti30
Fiber tract segmentation was already done for subject /Users/jyeatman/git/AFQ/data/control_02/dti30
Fiber tract cleaning was already done for subject /Users/jyeatman/git/AFQ/data/control_02/dti30
Computing Tract Profiles for subject /Users/jyeatman/git/AFQ/data/control_02/dti30
Whole-brain tractography was already done for subject /Users/jyeatman/git/AFQ/data/control_03/dti30
Fiber tract segmentation was already done for subject /Users/jyeatman/git/AFQ/data/control_03/dti30
Fiber tract cleaning was already done for subject /Users/jyeatman/git/AFQ/data/control_03/dti30
Computing Tract Profiles for subject /Users/jyeatman/git/AFQ/data/control_03/dti30
Run a t-test to compare FA along each fiber tract for patients vs. controls
for jj = 1:20
[h(jj,:),p(jj,:),~,Tstats(jj)] = ttest2(afq.patient_data(jj).FA,afq.control_data(jj).FA);
end
Make Tract Profiles of T statistics
fg = dtiReadFibers(fullfile(sub_dirs{3},'fibers','MoriGroups_clean_D5_L4.mat'));
dt = dtiLoadDt6(fullfile(sub_dirs{3},'dt6.mat'));
numNodes = 100;
[fa, md, rd, ad, cl, volume, TractProfile] = AFQ_ComputeTractProperties(fg,dt,numNodes);
for jj = 1:20
TractProfile(jj) = AFQ_TractProfileSet(TractProfile(jj),'vals','pval',p(jj,:));
TractProfile(jj) = AFQ_TractProfileSet(TractProfile(jj),'vals','Tstat',Tstats(jj).tstat);
end
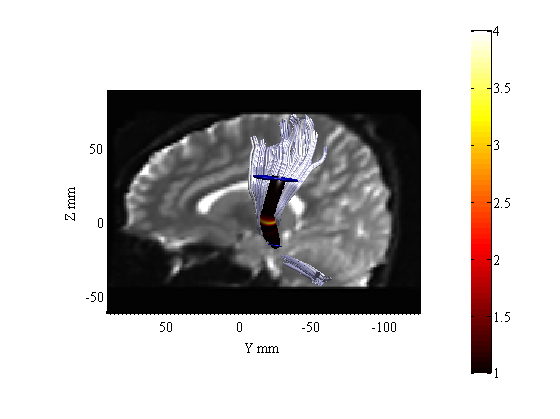
Render The tract Profiles of T statistics
cmap = 'hot';
crange = [1 4];
numfibers = 200;
AFQ_RenderFibers(fg(3),'color',[.8 .8 1],'tractprofile',TractProfile(3),...
'val','Tstat','numfibers',numfibers,'cmap',cmap,'crange',crange,...
'radius',[1 6]);
[roi1 roi2] = AFQ_LoadROIs(3,sub_dirs{3});
AFQ_RenderRoi(roi1,[0 0 .7]);
AFQ_RenderRoi(roi2,[0 0 .7]);
b0 = readFileNifti(dt.files.b0);
AFQ_AddImageTo3dPlot(b0,[-15,0,0]);
mesh can be rotated with arrow keys