Contents
Example AFQ analysis
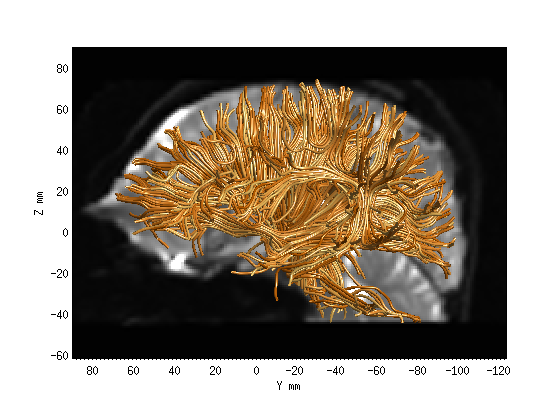
Step 1: Whole-brain tractography.
[AFQbase AFQdata AFQfunc AFQutil AFQdoc AFQgui] = AFQ_directories;
sub_dir = fullfile(AFQdata, 'control_01', 'dti30');
dt = dtiLoadDt6(fullfile(sub_dir,'dt6.mat'));
wholebrainFG = AFQ_WholebrainTractography(dt,'test');
AFQ_RenderFibers(wholebrainFG, 'numfibers',1000, 'color', [1 .6 .2]);
b0 = readFileNifti(fullfile(sub_dir,'bin','b0.nii.gz'));
AFQ_AddImageTo3dPlot(b0,[-2, 0, 0]);
scale=[2.0,2.0,2.0]mm, track=1, interp=1, step=1.0mm, fa=0.20, angle=30.0deg, puncture=0.20, minLength=50.0mm, maxLength=250.0mm
Tracking 33670 fibers (421 fibers per tick):
................................................................................
19833 fibers passed length threshold of 50.0 (out of 33670 seeds).
Elapsed time is 26.902815 seconds.
19833 fibers, mean length 97mm (max 251mm; min 50mm).
mesh can be rotated with arrow keys
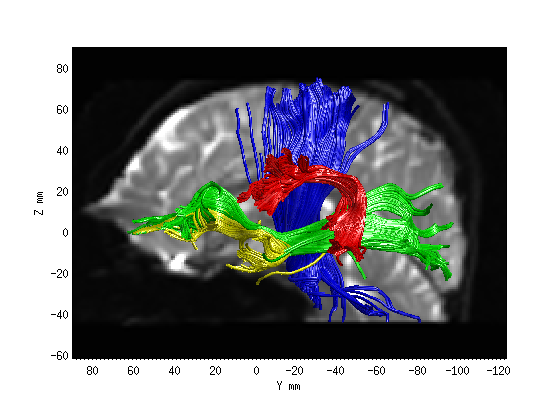
Step 2: Fiber tract segmentation
fg_classified = AFQ_SegmentFiberGroups(dt, wholebrainFG);
fg_classified = fg2Array(fg_classified);
AFQ_RenderFibers(fg_classified(3),'numfibers',400,'color',[0 0 1]);
AFQ_RenderFibers(fg_classified(11),'numfibers',400,'color',[0 1 0],'newfig',false)
AFQ_RenderFibers(fg_classified(17),'numfibers',400,'color',[1 1 0],'newfig',false)
AFQ_RenderFibers(fg_classified(19),'numfibers',400,'color',[1 0 0],'newfig',false)
AFQ_AddImageTo3dPlot(b0,[-2, 0, 0]);
You chose to recompute ROIs
Fibers that get as close to the ROIs as 2mm will become candidates for the Mori Groups
Smoothing by 0 & 8mm..
Coarse Affine Registration..
Fine Affine Registration..
3D CT Norm...
iteration 1: FWHM = 13.31 Var = 24.3369
iteration 2: FWHM = 10.26 Var = 2.3268
iteration 3: FWHM = 9.9 Var = 1.82368
iteration 4: FWHM = 9.811 Var = 1.68895
iteration 5: FWHM = 9.746 Var = 1.62451
iteration 6: FWHM = 9.729 Var = 1.60517
iteration 7: FWHM = 9.714 Var = 1.59288
iteration 8: FWHM = 9.708 Var = 1.5878
iteration 9: FWHM = 9.7 Var = 1.58208
iteration 10: FWHM = 9.701 Var = 1.58178
iteration 11: FWHM = 9.698 Var = 1.58025
iteration 12: FWHM = 9.698 Var = 1.57987
iteration 13: FWHM = 9.697 Var = 1.57926
iteration 14: FWHM = 9.697 Var = 1.57926
iteration 15: FWHM = 9.696 Var = 1.57881
iteration 16: FWHM = 9.697 Var = 1.57881
Computing inverse deformation...
dtiCleanFibers: Keeping 19727 out of 19833 fibers.
dtiSplitInterhemisphericFibers: Splitting every fiber below Z=-10
mesh can be rotated with arrow keys
ans =
[]
ans =
[]
ans =
[]
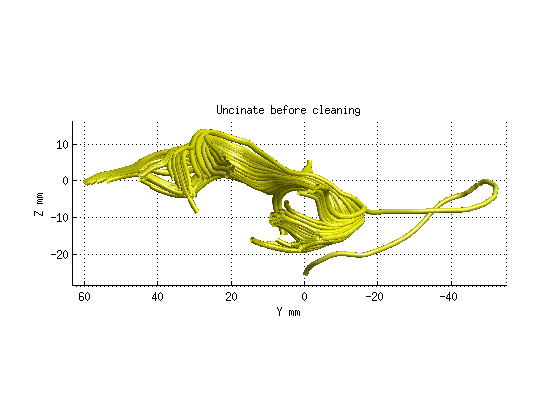
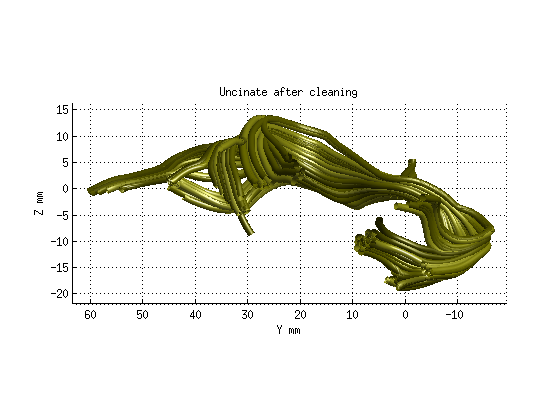
Step 3: Fiber tract cleaning
uf = fg_classified(17);
maxDist = 4;
maxLen = 4;
numNodes = 30;
M = 'mean';
maxIter = 1;
count = true;
uf_clean = AFQ_removeFiberOutliers(uf,maxDist,maxLen,numNodes,M,count,maxIter);
AFQ_RenderFibers(uf,'numfibers',1000,'color',[1 1 0]);
title('Uncinate before cleaning','fontsize',18)
AFQ_RenderFibers(uf_clean,'numfibers',1000,'color',[.5 .5 0]);
title('Uncinate after cleaning','fontsize',18)
for ii = 1:20
fg_clean(ii) = AFQ_removeFiberOutliers(fg_classified(ii),maxDist,maxLen,numNodes,M,count,maxIter);
end
Left Uncinate number of fibers: 110
Left Uncinate number of fibers: 106
mesh can be rotated with arrow keys
mesh can be rotated with arrow keys
Left Thalamic Radiation number of fibers: 260
Left Thalamic Radiation number of fibers: 234
Right Thalamic Radiation number of fibers: 220
Right Thalamic Radiation number of fibers: 202
Left Corticospinal number of fibers: 828
Left Corticospinal number of fibers: 777
Right Corticospinal number of fibers: 1031
Right Corticospinal number of fibers: 1009
Left Cingulum Cingulate number of fibers: 97
Left Cingulum Cingulate number of fibers: 86
Right Cingulum Cingulate number of fibers: 14
Left Cingulum Hippocampus number of fibers: 29
Left Cingulum Hippocampus number of fibers: 28
Right Cingulum Hippocampus number of fibers: 23
Right Cingulum Hippocampus number of fibers: 21
Callosum Forceps Major number of fibers: 262
Callosum Forceps Major number of fibers: 251
Callosum Forceps Minor number of fibers: 942
Callosum Forceps Minor number of fibers: 925
Left IFOF number of fibers: 300
Left IFOF number of fibers: 273
Right IFOF number of fibers: 165
Right IFOF number of fibers: 155
Left ILF number of fibers: 106
Left ILF number of fibers: 101
Right ILF number of fibers: 151
Right ILF number of fibers: 136
Left SLF number of fibers: 115
Left SLF number of fibers: 108
Right SLF number of fibers: 282
Right SLF number of fibers: 272
Left Uncinate number of fibers: 110
Left Uncinate number of fibers: 106
Right Uncinate number of fibers: 251
Right Uncinate number of fibers: 238
Left Arcuate number of fibers: 262
Left Arcuate number of fibers: 241
Right Arcuate number of fibers: 99
Right Arcuate number of fibers: 90
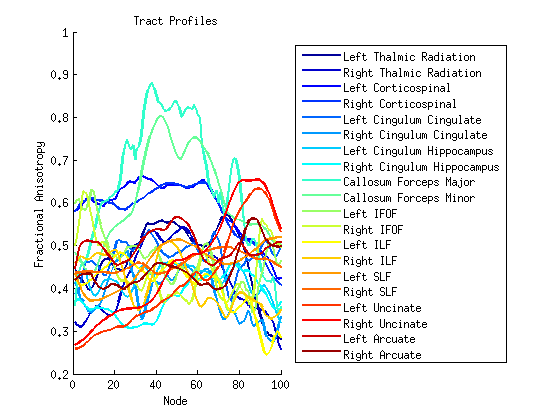
Step 4: Compute tract profiles
numNodes = 100;
[fa md rd ad] = AFQ_ComputeTractProperties(fg_clean, dt, numNodes);
figure; hold('on');
set(gca,'ColorOrder',jet(20));
plot(fa,'linewidth',2);
xlabel('Node');
ylabel('Fractional Anisotropy');
title('Tract Profiles');
fgNames={'Left Thalmic Radiation','Right Thalmic Radiation'...
'Left Corticospinal','Right Corticospinal', 'Left Cingulum Cingulate'...
'Right Cingulum Cingulate', 'Left Cingulum Hippocampus'...
'Right Cingulum Hippocampus', 'Callosum Forceps Major'...
'Callosum Forceps Minor', 'Left IFOF','Right IFOF','Left ILF'...
'Right ILF','Left SLF','Right SLF','Left Uncinate','Right Uncinate'...
'Left Arcuate','Right Arcuate'};
legend(fgNames,'Location','EastOutside' );
Step 5: Render Tract Profiles
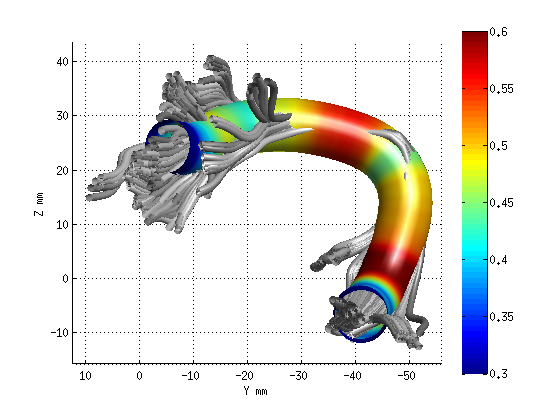
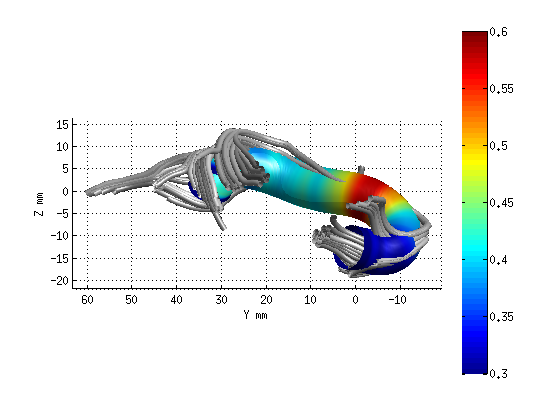
AFQ_RenderFibers(fg_clean(19),'dt',dt);
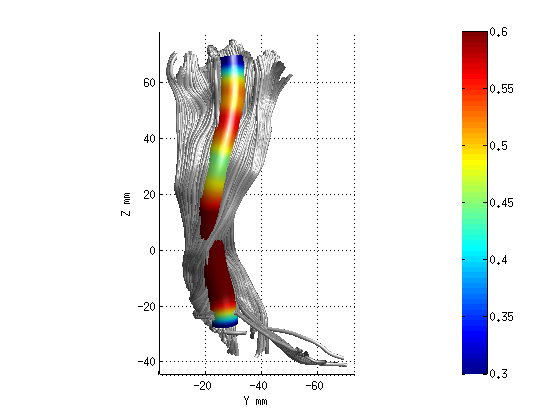
AFQ_RenderFibers(fg_clean(3),'dt',dt);
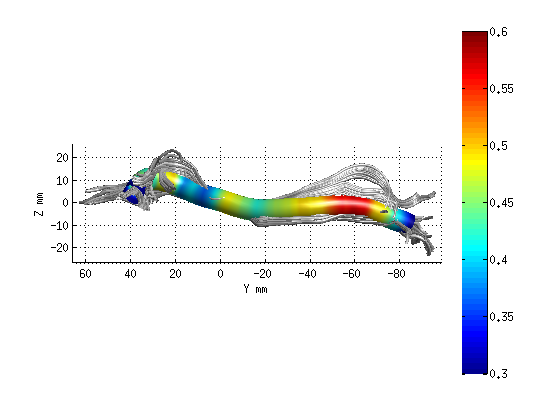
AFQ_RenderFibers(fg_clean(11),'dt',dt);
AFQ_RenderFibers(fg_clean(17),'dt',dt);
mesh can be rotated with arrow keys
mesh can be rotated with arrow keys
mesh can be rotated with arrow keys
mesh can be rotated with arrow keys